| Gene | Rap1gap |
| Chromosome | 4 |
| Deletion Start | 137,681,715 |
| Deletion End | 137,712,196 |
| Deletion Size | 30,482 |
| Genome Build | 38 |
|
|
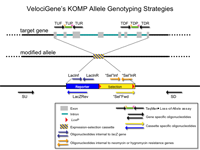
Primer Genomic Coordinates on Chromosome 4
Please be aware that these oligonucleotides have been validated only by Taqman assay.
| Primer | Start | End | Sequence |
| TUF | 137,682,560 | 137,682,580 | TGTCAAGTGAGGCCAGCTTAG |
| TUP | 137,682,585 | 137,682,607 | TGTGGGACTTTCATCCAGGCAGC |
| TUR | 137,682,645 | 137,682,668 | GACTGGGAAAGCCAAGACTCATAG |
| TDF | 137,711,199 | 137,711,219 | TGCACATCTGAGCTAGACTGG |
| TDP | 137,711,226 | 137,711,250 | AGGGAGCCTCCCTTGTCCCTGGAAG |
| TDR | 137,711,274 | 137,711,293 | GCTCTGAGGTTCCCTGAATC |
PCR Primers
| Primer | Sequence |
| SU | TTTGCTGCTCTTCACACAGG |
| SD | GCTCACTATTGGCTCACATG |
| LacInR | TTGACTGTAGCGGCTGATGTTG |
| LacInf | GGTAAACTGGCTCGGATTAGGG |
| NeoInR | TACTTTCTCGGCAGGAGCAAGGTG |
| NeoInF | TTCGGCTATGACTGGGCACAACAG |
| LacInZRev | GTCTGTCCTAGCTTCCTCACTG |
| NeoFwd | TCATTCTCAGTATTGTTTTGCC |
Predicted PCR Products
| Forward Primer | Reverse Primer | Length | WT | HET | KO |
| SU | LacZRev | bp | - | + | + |
| TUF | TUR | 109 bp | + | + | - |
| TDF | TDR | 95 bp | + | + | - |
| NeoFwd | SD | bp | - | + | + |
| LacInf | LacInR | 210 bp | - | + | + |
| NeoInf | NeoInR | 282 bp | - | + | + |
Available Clones
ES Cell ordering information can be found at the
KOMP Repository.
| Clone Name | Parental ES Cell | Mouse Strain | Cassette Copy Number |
| 12246A-F9 | VGB6 | C57BL/6NTac | |
| 12246A-H4 | VGB6 | C57BL/6NTac | |