| Gene | Acsm2 |
| Chromosome | 7 |
| Deletion Start | 119,563,508 |
| Deletion End | 119,596,056 |
| Deletion Size | 32,549 |
| Genome Build | 38 |
|
|
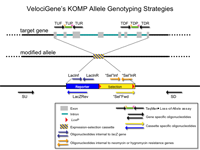
Primer Genomic Coordinates on Chromosome 7
Please be aware that these oligonucleotides have been validated only by Taqman assay.
| Primer | Start | End | Sequence |
| TUF | 119,563,666 | 119,563,686 | GGCCAGCGTAGAGAAGGTAAG |
| TUP | 119,563,692 | 119,563,721 | ACTGGATGGAACAGCTTGAATAAGGTCACA |
| TUR | 119,563,767 | 119,563,787 | GACCGTCCTGTTCACAGTTTC |
| TDF | 119,595,637 | 119,595,655 | TCTGGGCCATGTCGTGTTC |
| TDP | 119,595,660 | 119,595,688 | TGCTGCTTCTGTTCTCTTCATGACCATGT |
| TDR | 119,595,729 | 119,595,748 | ACCAGGGCAGGAAGCAGTTG |
PCR Primers
| Primer | Sequence |
| SU | TTTCTGGGATATGCGGTCTC |
| SD | TAGGACATGGGCATGACAAC |
| LacInR | TTGACTGTAGCGGCTGATGTTG |
| LacInf | GGTAAACTGGCTCGGATTAGGG |
| NeoInR | TACTTTCTCGGCAGGAGCAAGGTG |
| NeoInF | TTCGGCTATGACTGGGCACAACAG |
| LacInZRev | GTCTGTCCTAGCTTCCTCACTG |
| NeoFwd | TCATTCTCAGTATTGTTTTGCC |
Predicted PCR Products
| Forward Primer | Reverse Primer | Length | WT | HET | KO |
| SU | LacZRev | bp | - | + | + |
| TUF | TUR | 122 bp | + | + | - |
| TDF | TDR | 112 bp | + | + | - |
| NeoFwd | SD | bp | - | + | + |
| LacInf | LacInR | 210 bp | - | + | + |
| NeoInf | NeoInR | 282 bp | - | + | + |
Available Clones
ES Cell ordering information can be found at the
KOMP Repository.
| Clone Name | Parental ES Cell | Mouse Strain | Cassette Copy Number |
| 12702A-D11 | VGB6 | C57BL/6NTac | |
| 12702A-G2 | VGB6 | C57BL/6NTac | |
| 12702A-D12 | VGB6 | C57BL/6NTac | |