| Gene | C1qa |
| Chromosome | 4 |
| Deletion Start | 136,897,834 |
| Deletion End | 136,896,139 |
| Deletion Size | 1,696 |
| Genome Build | 38 |
|
|
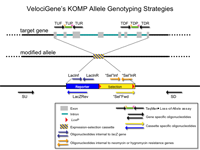
Primer Genomic Coordinates on Chromosome 4
Please be aware that these oligonucleotides have been validated only by Taqman assay.
| Primer | Start | End | Sequence |
| TUF | 136,897,275 | 136,897,299 | GAGAAAGATCAGCCAGTATCAACAG |
| TUP | 136,897,252 | 136,897,274 | TCACTTGCATGTCCTTCCAGCCC |
| TUR | 136,897,212 | 136,897,232 | TAACAGGGTAGAGTGGACAGG |
| TDF | 136,896,436 | 136,896,453 | CACACGGGTCGCTTCATC |
| TDP | 136,896,412 | 136,896,435 | TGTGCAGTGCCCGGCTTCTATTAC |
| TDR | 136,896,381 | 136,896,402 | GGTCCCACTTGGAGATCACTTG |
PCR Primers
| Primer | Sequence |
| SU | ATGCTGGACAGGTGCAGAG |
| SD | CACTGCACGGAAAGTCACAG |
| LacInR | TTGACTGTAGCGGCTGATGTTG |
| LacInf | GGTAAACTGGCTCGGATTAGGG |
| NeoInR | TACTTTCTCGGCAGGAGCAAGGTG |
| NeoInF | TTCGGCTATGACTGGGCACAACAG |
| LacInZRev | GTCTGTCCTAGCTTCCTCACTG |
| NeoFwd | TCATTCTCAGTATTGTTTTGCC |
Predicted PCR Products
| Forward Primer | Reverse Primer | Length | WT | HET | KO |
| SU | LacZRev | bp | - | + | + |
| TUF | TUR | 88 bp | + | + | - |
| TDF | TDR | 73 bp | + | + | - |
| NeoFwd | SD | bp | - | + | + |
| LacInf | LacInR | 210 bp | - | + | + |
| NeoInf | NeoInR | 282 bp | - | + | + |
Available Clones
ES Cell ordering information can be found at the
KOMP Repository.
| Clone Name | Parental ES Cell | Mouse Strain | Cassette Copy Number |
| 14732A-A1 | VGB6 | C57BL/6NTac | |
| 14732A-A12 | VGB6 | C57BL/6NTac | |
| 14732A-B12 | VGB6 | C57BL/6NTac | |
| 14732A-C2 | VGB6 | C57BL/6NTac | |
| 14732A-F7 | VGB6 | C57BL/6NTac | |
| 14732A-E3 | VGB6 | C57BL/6NTac | |